fig 3 from HICSS-26 pp674 1993
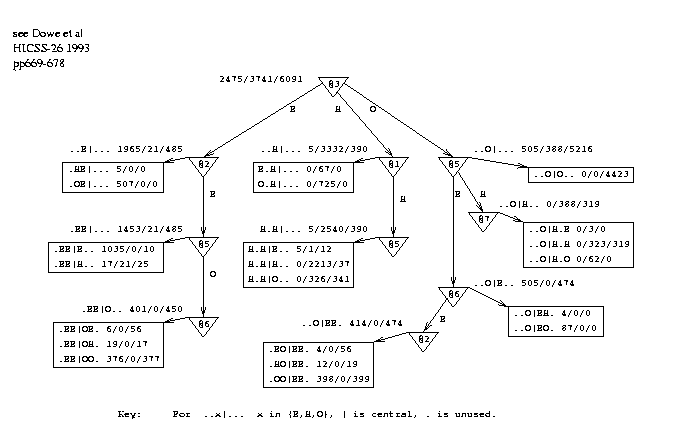
fig 4 from HICSS-26 pp674 1993
Partly supported by Australian Research Council grant A49030439.
Oliver and Wallace recently introduced the machine-learning technique of decision graphs, a generalisation of decision trees. Here it is applied to the prediction of protein secondary structure to infer a theory for this problem. The resulting decision graph provides both a prediction method and, perhaps more interestingly, an explanation for the problem. Many decision graphs are possible for the problem; a particular graph is just one theory or hypothesis of secondary structure formation. Minimum message length encoding is used to judge the quality of different theories and, for example, prevents learning the noise in the training data. Minimum message length encoding is a general technique in inductive inference.
The predictive accuracy for 3 states (Extended, Helix, Other) is in the range achieved by current methods. Many believe this is close to the limit for methods based on only local information. In addition, a decision graph is an explanation of a phenomenon. This contrasts with some other predictive techniques which are more "opaque". The reason for a prediction can be of more value than the prediction itself. Rules containing both conjunction and disjunction can be extracted from a decision graph in a natural way which makes for good expressive power. For example, they can express pair-wise interactions and conditional rules. Groups of related amino acids can be identified from the graph. The rules are probabilistic and indicate a degree of confidence in a prediction. The more confident predictions are generally more accurate. For this initial application of decision graphs to protein structure problems, the data comprised 10K amino acids from 50 proteins. The data was divided into various training and test sets.
decision graph, decision tree, explanation, machine learning, secondary structure.
A protein is a large and complex molecule consisting of one or more chains. Each chain consists of a back-bone, (N-Calpha-C)+, with one of the 20 possible amino-acids (table 1) attached to each alpha-carbon atom, Calpha. The 3-dimensional, folded shape of the protein is responsible for much of its biochemical activity and the prediction of this shape from the primary sequence of the protein is a major, long-term (!) goal of computing for molecular biology. There are several surveys of progress towards solving the problem[Ster83][Robs86][Tayl87][Reek88]. Techniques that have been applied include energy minimisation and molecular dynamics[Broo83], statistical methods[Garn78] and artificial intelligence methods[Lath87]. One line of attack is to use the different levels of protein structure that have been recognised[Schu88]:
It must be said that the boundaries between some of these levels can be blurred.
Today it is relatively easy to find the primary sequence of a protein but the full 3-D structure can only be obtained by x-ray crystallography which is both difficult and time-consuming. Many primary sequences but relatively few 3-D structures are known. Certain local or secondary structures recur in proteins: helices, turns, and extended strands which can form beta-sheets. Reliably predicting secondary structure from primary sequence would reduce the 3-D prediction problem from arranging or packing hundreds of amino-acids to packing tens of secondary structures. This explains the considerable interest in secondary structure prediction. Current methods achieve success rates in the 55%[Kabs83b] to 65%[Garn91] range for 3 states - extended (E), helix(H) and other(O) - on arbitrary proteins. Many believe this to be close to the limit for methods based only on local information or interactions. The remaining amount is thought to be due to long-range factors. It should also be noted that restricting the type of proteins, or using other information, for example the known structure of a protein with a similar primary sequence, if there is one, can much improve predictions.
1 A Alanine Ala 2 C Cysteine Cys 3 D Aspartate Asp 4 E Glutamate Glu 5 F Phenylalanine Phe 6 G Glycine Gly 7 H Histidine His 8 I Isoleucine Ile 9 K Lysine Lys 10 L Leucine Leu 11 M Methionine Met 12 N Asparagine Asn 13 P Proline Pro 14 Q Glutamine Gln 15 R Arginine Arg 16 S Serine Ser 17 T Threonine Thr 18 V Valine Val 19 W Tryptophan Trp 20 Y Tyrosine Tyr Table 1: Amino Acid Codes
We use a machine-learning program to infer a decision graph[Oliv91] for predicting secondary structure from primary sequence. The training data consists of windows from proteins of known structure. The central amino-acid in a window is assigned a secondary structure state (E,O,H) by an external expert in the form of the DSSP program[Kabs83a]. The decision graph embodies rules that have been inferred from the training data, to assign states. The graph or the rules can then be used to predict states for new proteins. Decision graphs are a generalisation of the well-known decision trees[Quin89]. Graphs have certain advantages over trees for this problem, see section 2. Logical rules containing conjunction and disjunction are naturally derived from a graph. The rules are probabilistic and indicate a degree of confidence in a prediction. The more confident predictions tend to be more accurate. The graph is a theory or hypothesis about structure prediction. The rules themselves can be taken as an explanation and can be as important as the predictions that they make. A second graph is used to model the serial correlation in secondary structure and to smooth the predictions of the first graph.
Many decision graphs are possible for the secondary structure problem. Minimum message length (MML) encoding is used as the criterion to guide the search for a good one. A particular graph is (just) a theory or hypothesis. A good graph will predict the data well but a complex graph might be fitting the idiosyncrasies[Holl89] or noise in the training data. Under the MML principle, there is a larger overhead for a complex graph than for a simple one - it must pay its way. This is a computational version of Occam's razor and balances the trade-off in graph complexity. It is described in more detail in section 3.
We believe that the MML criterion is the best yardstick on which to compare methods, whether based on information theory or not, but it is not common practice and so we give the percentage of correct predictions for comparison purposes. Initial experiments used a database of 50 non-homologous proteins and a window of size 7. Test sets consisted of individual proteins and of sets of 10 proteins, the remaining proteins being used as training data. The average success rate for 3 states was 44% to 63%, averaging 54%, which is in the typical range, at the lower end.
Many approaches have been taken to computer prediction of secondary structure. There are reviews by Kabsch and Sander[Kabs83b] in 1983, Schulz[Schu88] in 1988 and Garnier and Levin[Garn91] in 1991. Some of the difficulties in comparing different methods are that standard data-sets are rarely used and that entries in the structure database are "improved" and even deleted.
The Chou-Fasman method[Chou74] is one of the earliest methods
and was initially carried out by hand.
It is based on coefficients for each pair
Garnier, Osguthorpe and Robson defined a fully algorithmic
and objective method based on information theory
and now known as the GOR method[Garn78].
For each triple,
Neural-nets have been applied to secondary structure prediction[Holl89]. A typical neural-net fits a simple non-linear (sigmoidal) model to the data. The GOR method fits a linear model on -log odds-ratios, so one might expect a neural net to perform similarly, as is the case. On the other hand it is difficult to extract an explicit model from a neural-net to provide an explanation of how the predictions are made.
Muggleton et al[Mugg92a][Mugg92b] have applied Golem to secondary structure prediction. Golem is a general system to infer rules from examples. The rules are in Horn-clause form, essentially a Prolog program. These rules must express disjunction indirectly but otherwise are similar in general form to those created from decision graphs. Unfortunately, the application of Golem was to a restricted set of alpha domain type proteins, making direct comparison with other methods difficult.
The consensus is that the GOR and several other methods can achieve 60-odd% accuracy on arbitrary proteins while the methods of Chou and Fasman and of Lim are less accurate[Garn91]. Higher accuracy is possible in restricted sets of proteins or by using other information. The GOR method is based on a simple, explicit, probabilistic model. Logical rules, created by hand or as inferred by Golem or derived from a decision graph, are in a general or universal language and can be said to be more of an explanation in the everyday sense.
Oliver and Wallace[Oliv91] recently introduced the notion of a decision graph which is a generalisation of the well-known decision tree[Quin89]. A decision function is a mapping from a vector of attributes of a "thing" to the class of the thing, eg. from the amino acids within a window to the state of the central amino acid in the window. Decision trees and graphs represent the same class of decision functions but graphs are more efficient in learning rules with a disjunctive nature. This can variously be taken to mean that (for such cases): a graph is learned more quickly, graphs are more expressive or a graph is a more succinct theory.
A decision tree (figure 1) contains decision nodes and leaves. A decision node specifies a test on an attribute of a thing. The test can be a k-way split on a discrete value or can be an inequality on a real value. A leaf node assigns a class to things. The assignment can be probabilistic[Wall92] and indicate a degree of confidence. To use a tree given a thing, a path is traced from the root of the tree to a leaf following arcs as indicated by the results of tests. A path in a tree represents a conjunction of tests: p&q&r&... .
function to be learned:
class #1 = (@1 & @2) or (@3 & @4)
class #2 = ~ #1
binary attributes @1, @2 @3 and @4.
@1
. .
t. .f
. .
@2 @3
. . . .
t. .f t. .f
. . . .
#1 @3 @4 #2
. . . .
t. .f t. .f
. . . .
@4 #2 #1 #2
. .
t. .f
. .
#1 #2
Figure 1: Example Decision Tree
A decision graph (figure 2) can contain join nodes
in addition to decision nodes and leaves.
A join node represents the disjunction of the paths
leading to it:
function to be learned:
class #1 = (@1 & @2) or (@3 & @4)
class #2 = ~ #1
@1
. .
t. .f
. .
@2 .
. . .
t. .f .
. ..
#1 join
.
.
@3
. .
t. .f
. .
@4 #2
. .
t. .f
. .
#1 #2
Figure 2: Example Decision Graph
The most obvious attributes in secondary structure prediction are the amino acids in a window but 20 is a very high arity or fan-out for an attribute. A decision node divides the training data into 20 subsets. 2 levels of decision nodes divide it into 400 subsets. This rapidly halts progress in the inference of a decision tree. On the other hand, decision graphs can delay this problem by joining paths with related outcomes, by adding the data flowing along these paths, and learning a richer theory. Note, it is also possible to add redundant attributes, possibly with lower arity such as classes that amino acids belong to, but that is not considered here.
There are a great many possible decision graphs for secondary structure prediction. Minimum message length (MML) encoding is used to evaluate graphs and to guide the search for a "good" one. The search can be difficult and lookahead and heuristics are necessary. Global optimality cannot generally be guaranteed in feasible time. However, any two graphs can be compared with confidence.
Minimum message length (MML) encoding is a general information-theoretic principle[Chai66]. Given data D we wish to infer a hypothesis H. In general there are many possible hypotheses and we can only speak of better or worse hypotheses. Imagine a transmitter who must send a message containing the data to a receiver who is a considerable distance away. If the transmitter can infer a good hypothesis it can be used to compress the data. However any details of the hypothesis that the receiver does not already know, ie. that are not common knowledge, must be included in the message or it will not be comprehensible. The message paradigm keeps us honest - nothing can be passed under the table. We therefore consider two-part messages, which transmit first a hypothesis H and then transmit D using a code which would be optimal if the hypothesis were true. From information-theory, the message length (ML) in an optimal code of an event E of probability P(E) is -log2(P(E)) bits. Therefore the best hypothesis is the one giving the shortest total message.
ML(E) = -log2(P(E)), event E of probability P(E). Bayes' theorem: P(H&D)= P(D).P(H|D) = P(H).P(D|H) ML(H&D)=ML(D)+ML(H|D)=ML(H)+ML(D|H) posterior log odds-ratio: -log2(P(H|D)/P(H'|D)) = ML(H|D)-ML(H'|D) = (ML(H)+ML(D|H)) - (ML(H')+ML(D|H')) for hypotheses H and H', data D.
P(D|H) is the likelihood of the hypothesis - actually a function of the data given the hypothesis. P(H) is the prior probability of hypothesis H. Oliver and Wallace[Oliv91] discuss a prior for decision graphs. P(H|D) is the posterior probability of hypothesis H. The posterior odds-ratio of two hypotheses H and H' can be calculated from the difference in total message lengths: the better one gives the shortest message. The raw data itself constitutes a null-theory and gives a hypothesis test: any hypothesis that gives an overall message length longer than that of the null-theory is not acceptable.
The MML principle has been successfully applied to clustering or classification[Wall90], decision trees[Quin89][Wall92], decision graphs[Oliv91], string alignment[Alli92] and other problems.
Our present decision graphs are hypotheses of secondary structure formation. A graph makes probabilistic predictions of structure. If they are good predictions the states (E,O,H) can be transmitted very cheaply. Balancing this saving is the cost of transmitting the graph itself. Thus, simple and complex graphs are compared fairly. This prevents the fitting of noise in training data.
The data used in our experiments is a subset of the 75 proteins used by Garnier and Robson[Garn89, table XIII p449] and reported in 1989. That set does not contain homologous proteins. The use of standard data, even out of date data, would allow two or more methods to be directly compared. Unfortunately some entries have been superceded and removed and others may have been updated since 1989. The list used is given in table 2.
155c2 156b4 1abp4 1azu5 1bp21 1crn3 1ctx3 1ecd3 1est3 1fdx5 1gcn2 1gpd1 1hip5 1mbn2 1nxb2 1ovo1 1pcy3 1ppt2 1rei3 1rhd4 1rns5 1sbt3 1tim1 2act4 2alp5 2app1 2b5c1 2gch4 2hhb5 2mhb1 2pab3 2rhe3 2sga5 2sns2 2sod2 2ssi5 351c5 3c2c4 3cna2 3cyt4 3dfr2 3fxc1 3fxn1 3pgm4 3tln4 4pti4 4rxn5 5cpa1 7cat2 7lyz3 Test sets: 1:10test1 2:10test2 3:10test3 4:10test4 5:10test5 also 1gpd, 2hhb, 7cat. Training set is the complement of the test set in each case. Table 2: Proteins Used
The DSSP program[Kabs83a] was used to assign secondary structure states to known 3-D structures. A window of size 7 was used in the main experiments. This is sufficient to give reasonable predictions and to infer a rich graph and one that can be presented on a page. Some trials have been done with larger and smaller window sizes. The window was slid along each protein, the amino acids in the window becoming the attributes, and the state becoming the class, of a "thing". Any window where DSSP reported an error, an exception or a chain break was discarded, principally because it was not always clear what other methods did in such cases. This left 8.5K windows in all.
In order to test the predictive power of graphs, the data was divided randomly into 5 groups of ten proteins: 10test1 to 10test5. Each group served as test data for a graph inferred from the union of the remaining four groups. In addition, the longest protein was used as test data for a graph inferred from all the remaining proteins. This was also done for the second and fourth longest proteins. This is perhaps the severest and least biased kind of test of a predictive method - testing on arbitrary proteins when trained on non-homologous proteins.
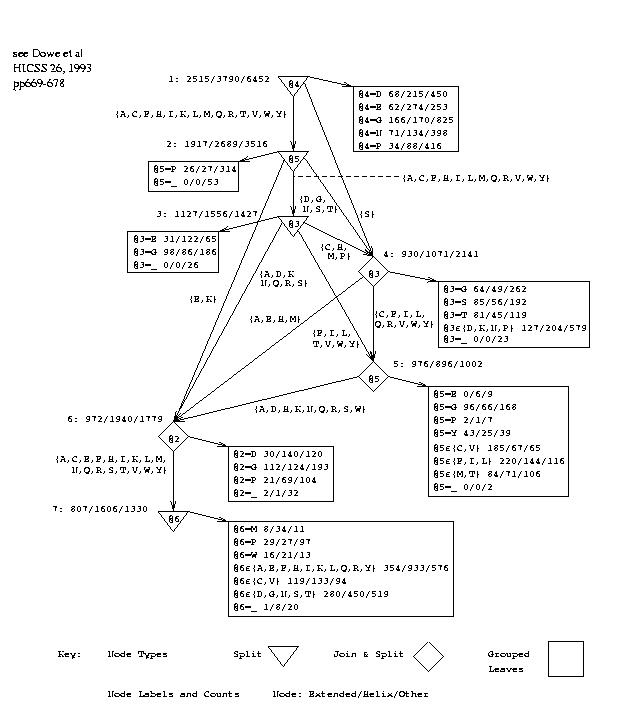
Figure 3 shows the decision graph inferred from the training set corresponding to test set 10test3, using a window size of 7.
fig 3 near hereFigure 3: Decision Graph with Window Size 7
Attributes or positions are numbered 1 to 7 by the program (rather than -3 to +3), the central, predicted amino acid being at position 4, and this numbering is used in the discussion. Rectangular boxes represent leaves. The observed frequencies of the states, E/O/H, are given in the leaves and at certain other points in the graph. The frequencies give estimates of the probability of each state[Wall92]. Study of the graph reveals various groups of amino acids. These groups were formed automatically by the program and were not provided as background knowledge. A further possibility to be investigated is to provide these or other groupings in the data to the program as background knowledge.
The graphs inferred for different data sets vary slightly. It is a close thing whether it is better for the top-level test to be on the central amino acid (4) or on the following one (5), as it is in this graph. The reason is probably this: The central amino acid is a major factor but proline (P) in the 5th position is a strong indicator of turns[Robs76] and in this graph leads immediately to a leaf that strongly predicts `other', `O', in 270/310 cases. We also note more tests in the graph that refer to proline and lead to leaves that are strongly `other'.
The top-level test also leads into join nodes, or disjunctions, which form three groups of amino acids:
G1 = [AEKQ], G2 = [FILV], G3 = [CDGHMNRSTWY].
The set of values in G1 is then split on position 4, where values in G1.1 = [AEKQR] lead to a split on position 3, where values in G1.1.1=[AELMQV] lead to a leaf making a strong helix prediction:
if Att3 in [AELMQV] & Att4 in [AEKQR] & Att5 in [AEKQ] then P(State is Helix) ~ 117/170
A, E, K and Q appear in at least two of G1, G1.1 and G1.1.1. A and E are known to be strong indicators of helices[Schu88], and K often appears at the C-terminal end of helices[Robs76]. Note, K is in the groups for positions 4 and 5 but not 3. EKQ are close to each other, with A not far away, on the codon ring defined by Swanson[Swan84, p188, fig 1], where proximity indicates similarity of codons in the genetic code and similarity of physico-chemical properties.
Robson[Robs76] performed a cluster analysis on amino acids based on secondary structure preferences and reported, "The best defined group is composed of non-polar residues. ... [FILMV]". This is no other than G2+[M]. FILV and M occur next to each other in the codon ring[Swan84] and ILV form Taylor's aliphatic group[Tayl86, p210, fig 3]. These amino acids tend to form extended strands. If position 5 is in G2, the arc leads to a join node and then a split on position 3 and interestingly not on position 4 although it is joined with descendants of a split on position 4 with values [FQVY]. Tracking descendants where position 3 is non-polar we find
if Att3 in [FTY] & (Att5 in [FILV] or ...) then P(State is Extended) ~ 120/292 if Att3 in [IV] & (Att5 in [FILV] or ... ) then P(State is Extended) ~ 61/104
Position 3 being in [LM] leads to a further split on position 2.
Jimenez-Montano examined various classifications of the amino acids. G1 and G2 fit most naturally with a hierarchy[Jimi84, p653, fig 3] derived from Lim's work[Lim74] on hand-crafted rules. G2+[M] is contained in one leaf of that classification and most of G1.1, [EKQR], is contained in a second leaf. G2+[M] also forms most of the leaves of a subtree in Jimenez-Montano's own favoured hierarchy[Jimi84, p649, fig 1].
The group G3 might appear to be a "miscellaneous" collection but a rather similar group G4 = [CDGHIKNPRSTWY] appears in
if (Att3 in G4 & ((Att4 in G1.1 & Att5 in G1) or ... ) or (Att5 in G3 & Att4 in [EHKR]) or ... then P(State is not Extended) ~ 1657/1807
It is evident that members of G3 intersection G4 = [CDGHNRSTWY] are strongly anti-extended in certain contexts.
The above decision graph for secondary structure prediction does not model the serial correlation in secondary structure. It may be possible to extend the theory of decision trees and graphs to model series directly, and some alternatives are being investigated, but currently the initial predictions are smoothed. This smoothing is carried out by a second decision graph which models just the serial correlation in secondary structure. It improves the success rate by about 3.5%.
Figure 4 shows a smoothing graph derived from all 50 proteins. Note that the smoothing graph used in a test was inferred only from the appropriate training data. The states of the 6 flanking amino acids, positions 1 to 3 and 5 to 7, are the "attributes" of a "thing" and the state of the central amino acid (4) is the "class". When the graph is used, the predicted states of the flanking positions are used as inputs. (A further possibility would be to use the 7 predicted states as the attributes when inferring the smoothing graph.) Each leaf in the graph has pattern(s) associated with it that correspond to the path(s) to the leaf. Note that the 2 patterns associated with leaf `W' are reversals of each other. Similarly, reversed pairs of patterns are associated with leaves `X', `Y' and `Z'. In this way, the graph reveals symmetries that would be hard to express in a decision tree.
figure 4 near hereFigure 4: Decision Graph Demonstrating Serial Correlation in Conformations
The question arises of how to reconcile the possibly conflicting results of the prediction graph `PG' and the smoothing graph `SG'. The following procedure was used to make a smoothed sequence of predictions for a test protein. Any given sequence of predictions for the protein can be given a probability under PG and another probability under SG. To do this for one of the graphs, the probabilities of the predicted states can be found - from the leaves of the graph - and multiplied together. Equivalently, the -log probabilities are taken and added, which gives an empirical energy function for the sequence of predictions under the graph. This indicates how consistent the predictions are with the graph. The energy is calculated for both PG and SG and the results added with a parameter to control the relative weight given to each graph. This ratio was optimised on the training set, typically being approximately 10:1 as the unsmoothed predictions were fairly sound. The search for a sequence of predictions was carried out by simulated annealing[Kirk83], starting from either the unsmoothed predictions or random predictions.
unsmoothed smoothed
test tree graph graph
set Train Test Train Test Train Test
------- ---------- ---------- ----------
10test1 59.9 47.2 58.4 48.7 62.9 53.2
10test2 58.1 51.2 53.6 50.3 59.0 55.5
10test3 62.9 46.3 56.8 54.3* 63.0 55.0
10test4 64.0 49.2 56.8 51.8 62.0 54.5
10test5 58.6 45.8 60.6 47.0 68.3 51.3
---- ----
average 50.4 53.9
* see figure 3
1gpd 60.8 49.8 55.4 55.0 60.9 62.8
2hhb 56.6 39.3 55.7 47.6 60.2 44.0
7cat 62.3 52.6 61.1 53.7 65.2 56.7
---- ----
average 52.1 54.5
Table 3: Prediction Success Rates (EOH)
The numerical results of the main experiments are summarised in table 3. This gives the success rates for 3 states (EOH) for the prediction graph alone and for the smoothed results, on the training data and on the test data. The smoothing increases rates by about 3.5%. Results for a decision tree are also included for comparison.
A decision graph makes probabilistic predictions. A leaf node indicates a degree of confidence in a prediction. Some leaves are ambivalent - they effectively make guesses - and perhaps their predictions should be discounted. For example, if any prediction where the most probable state has probability less than 0.5 is ignored, the unsmoothed success rate on 10test3 rises from 54.3% to 57.0% although the coverage falls from 100% to 60%.
A decision graph is a theory or explanation of secondary structure prediction. Logical rules containing conjunction and disjunction were extracted from it. For a database of 50 proteins and a window of 7, the success rate is in the 44% to 63% range, averaging 54%, at the lower end of the range for current methods. Future work includes giving the decision graph program some background knowledge in terms of amino acid classes, trying both larger databases and restricted classes of proteins, using different window sizes, using a larger number of secondary structure states and investigating different smoothing methods.


Also see: [Bioinformatics].